Sie befinden sich hier
Inhalt
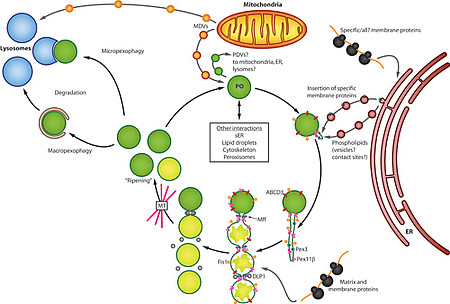
Peroxisomes are ubiquitous organelles involved in a variety of metabolic pathways such as fatty acid β-oxidation, etherlipid synthesis or detoxification of reactive oxygen species (Islinger et al. 2011, Islinger et al. 2012). Along with this interconnection with numerous cellular pathways peroxisomes have to react upon numerous internal and external stimuli in order to modulate their abundance, protein content and interaction with other subcellular compartments (Schrader et al. 2012, Fig. 1).
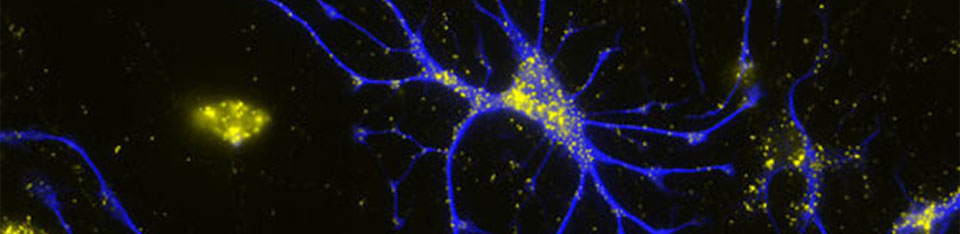
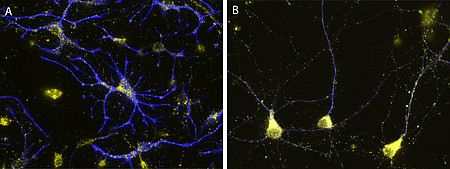
Their importance for human health and disease is documented by the existence of several inherited peroxisomal disorders, which have detrimental symptoms such as neonatal hypotonia, craniofacial dysmorphy, neuronal myelination and migration defects, seizures, hepatomegaly, liver cirrhosis, and renal cysts generally leading to death in early childhood. Furthermore peroxisomal alterations have been reported in neurodegenerative disorders such as Alzheimer’s disease (Kou et al. 2011, Lizard et al. 2012). Despite the obvious neurological phenotype of peroxisomal genetic disorders our current knowledge on the role of these organelles in the brain is comparably scarce. Whereas the detrimental effects of peroxisomes in the brain have been mainly linked to the neuron nurturing function of oligodendrocytes, peroxisomes can be also observed in considerable amounts in neurons and astrocytes (Fig. 2), where their role is still more enigmatic.
In this regard our research interests in peroxisomes biology focuses on a principle characterization of the peroxisomal functions in neurons and glial cells of the brain to increase our understanding of their contribution to human health and disease in this organ of outstanding importance. With respect to this intention our current experimental strategy is divided into two main research lines:
Project 1: Characterization of the peroxisomal brain proteome
Our current knowledge about the protein composition of peroxisomes is largely based on the acquisition of the proteome of the organelles isolated from the liver and kidney (Wiese et al. 2007, Islinger et al. 2007). However, an isolation procedure to enrich peroxisomes from the brain to a purity sufficient for a subsequent mass spectrometrical characterization is at the moment not available. To solve this problem we are presently developing a purification procedure combining traditional and inventive separation techniques such as density gradient centrifugation and free flow electrophoresis.
Peroxisome enriched and depleted fractions will be subsequently compared by quantitative mass spectrometry in order to identify bona fide peroxisomal proteins. Newly identified brain-specific peroxisomal proteins will be further characterized by expression/knockdown in neuronal primary cultures from different brain areas. (taken from Islinger et al. 2012)
Project 2: Plasticity of peroxisomes in the brain
Peroxisomes have to adapt their abundance and enzyme content according to the requirements of the overall homeostasis of the cell. In liver and kidney p the peroxisome proliferator activated receptor a (PPARα) induces organellar proliferation and increased expression of peroxisomal β-oxidation enzymes. In the brain, however, PPARα is only very moderately expressed suggesting that other regulation networks are involved in the control of organelle activity and proliferation.
To decipher new factors and pathways triggering peroxisome proliferation in neurons and glia we apply a screening approach using primary cerebellar cultures to monitor changes in peroxisomes numbers upon administration of metabolites, hormones and growth factors into the culture medium. Further we are interested in characterizing the function of proteins involved in the induction process and to assess the cellular biochemical alterations induced by the organelle’s proliferation.
Publications
Delille HK, Agricola B, Guimaraes SC, Borta H, Lüers GH, Fransen M, Schrader M. 2010. Pex11pbeta-mediated growth and division of mammalian peroxisomes follows a maturation pathway. J Cell Sci. 123, 2750-2762. doi: 10.1242/jcs.062109
Islinger M, Schrader M. 2011. Peroxisomes. Curr Biol. 21, R800-801. doi: 10.1016/j.cub.2011.07.024
Islinger M, Grille S, Fahimi HD, Schrader M. 2012. The peroxisome: an update on mysteries. Histochem Cell Biol. 137, 547-574. doi: 10.1007/s00418-012-0941-4
Islinger M, Lüers GH, Li KW, Loos M, Völkl A. 2007. Rat liver peroxisomes after fibrate treatment. A survey using quantitative mass spectrometry. J Biol Chem. 282, 23055-23069.
Kou J, Kovacs GG, Höftberger R, Kulik W, Brodde A, Forss-Petter S, Hönigschnabl S, Gleiss A, Brügger B, Wanders R, Just W, Budka H, Jungwirth S, Fischer P, Berger J. 2011. Peroxisomal alterations in Alzheimer's disease. Acta Neuropathol. 122, 271-283. doi: 10.1007/s00401-011-0836-9
Lizard G, Rouaud O, Demarquoy J, Cherkaoui-Malki M, Iuliano L. 2012. Potential roles of peroxisomes in Alzheimer's disease and in dementia of the Alzheimer's type. J Alzheimers Dis. 29, 241-254. doi: 10.3233/JAD-2011-111163
Schrader M, Bonekamp NA, Islinger M. 2012. Fission and proliferation of peroxisomes. Biochim Biophys Acta. 1822, 1343-1357. doi: 10.1016/j.bbadis.2011.12.014
Wiese S, Gronemeyer T, Ofman R, Kunze M, Grou CP, Almeida JA, Eisenacher M, Stephan C, Hayen H, Schollenberger L, Korosec T, Waterham HR, Schliebs W, Erdmann R, Berger J, Meyer HE, Just W, Azevedo JE, Wanders RJ, Warscheid B. 2007. Proteomics characterization of mouse kidney peroxisomes by tandem mass spectrometry and protein correlation profiling. Mol Cell Proteomics. 6, 2045-2057.
Kontextspalte
Group Leader

PD Dr. Markus Islinger
Phone +49 621 383-6898
markus.islinger@medma.uni-heidelberg.de